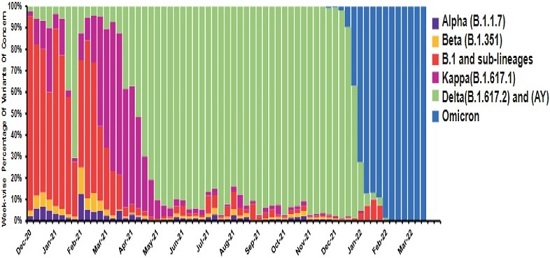
Figure Legend: The bar graph represents distribution of different variants from December 2020 to March-2022.
Different SARS-CoV-2 variants drove three major COVID-19 waves in India. A group of researchers has studied the effects of SARS-COV-2’s Variants of Concern (VOC) using genomic surveillance.
The study is based on the surveillance and sequenced samples in Pune during December 2020 to March 2022.
“The next-generation technology was employed to sequence SARS-CoV-2 genomes obtained from RNA samples of COVID-19 positive patients (by RT-PCR). For our study, we considered 10,496 samples which were sequenced using two different platforms at IISER and NCL, Pune,” informs Dr Krishanpal Karmodiya, the lead researcher.
A city-wide network of researchers, clinicians, and pathology diagnostic laboratories was formed for genome surveillance of COVID-19 in Pune. The sequencing data indicated that different VOCs such as Delta, Kappa, and Omicron were predominantly driving the second and third waves in Pune.
The Covid-seq team at IISER Pune (from Left to Right) – Dr Soumen Khan, Dr Dipti Deshpande, Ms. Nikita Shah, Ms. Unnati Bhalerao, Ms. Manisha Tupekar, Ms. Rutuja Sawant, Ms. Pradnya Kadam, Dr Aurnab Ghose and Dr Krishanpal Karmodiya
The team considered various factors for studying the overall effect of these variants which included infectivity rate considering age and gender.
“The age group with the highest number of infections was from 31 to 45 years during the phase when Delta variant was prevalent and age group 16-30 years during the Omicron variant. However, there was no significant difference observed in infectivity rate considering gender,” adds Dr Karmodiya.
Mutations in different variants were also compared; and among 10,496 whole genome sequences, 98 unique single mutations were detected on the Spike protein which plays an important role in initiation of the infection process.
The ‘band of five’ outbreak data analytics approach, integrating five different types of data, highlights the importance of a strong surveillance system for understanding the spatiotemporal evolution of the SARS-CoV-2 genome in Pune. “The findings have important implications for pandemic preparedness and could be critical tools for understanding and responding to future outbreaks,” the researchers believe.
The work was supported by the Rockefeller Foundation, CSIR, Villoo Poonawalla Foundation (VPF), Janakidevi Bajaj Gram Vikas Sanstha (JBGVS), DST-Science and Engineering Research Board (SERB), DBT-Indian SARS-CoV-2 Genomics Consortium (INSACOG), and Johns Hopkins India Institute Covid Relief Fund.
The study has been published in the Journal of Infection and Public Health. The team comprised researchers from the CSIR-National Chemical Laboratory; the Indian Institute of Science Education and Research (IISER); The Pune Knowledge Cluster (PKC), Savitribai Phule Pune University, Pune; GenePath Diagnostics India Private Limited, Pune; and Byramjee Jeejeebhoy Government Medical College (BJGMC), Pune.
India Science Wire
ISW/SM/NCL/COVID-19/Eng/21/06/2023